[TOC]
数据集:Decathlon Task07_Pancreas
平台: Ubuntu为主 (windows作测试用,其中很多不准)
参考:
准备&Decathlon数据集使用
设定数据集路径
在环境路径中,设定网络的基本路径:
- [nnUNet_raw_data_base] - D:\Projects\nnUNet\data_base\nnUNet_raw_data
- [nnUNet_preprocessed] - D:\Projects\nnUNet\data_base\nnUNet_preprocessed
- [RESULTS_FOLDER] - D:\Projects\nnUNet\data_base\result_folder
| |
将Decathlon数据集进行转换
因为有一些数据是4D时序的数据,比如心脏,所以在这里,他将decathlon数据集转换成统一格式。
- 如果是3D数据,则文件内容不变,更改文件名,Eg: pancreas_001.nii.gz $\rightarrow$ pancreas_001_0000.nii.gz
- 如果是4D数据,则拆分成各个3D内容,在后面4位角标进行标注
| |
转换后,[nnUNet_raw_data_base]\ nnUNet_raw_data\ Task007_Pancreas 下的文件如下所示:
|-- nnUNet_preprocessed
|-- nnUNet_raw
| |-- nnUNet_cropped_data
| `-- nnUNet_raw_data
| `-- Task007_Pancreas
| |-- dataset.json
| |-- imagesTr
| | |-- pancreas_001_0000.nii.gz
| | |-- pancreas_004_0000.nii.gz
| | |-- ...
| | `-- pancreas_421_0000.nii.gz
| |-- imagesTs
| | |-- pancreas_002_0000.nii.gz
| | |-- pancreas_003_0000.nii.gz
| | |-- ...
| | `-- pancreas_420_0000.nii.gz
| `-- labelsTr
| |-- pancreas_001.nii.gz
| |-- pancreas_004.nii.gz
| |-- ...
| `-- pancreas_421.nii.gz
`-- result_folder
`-- nnUNet
进行前处理
| |
对数据进行整理(裁剪4D数据)
| |
在 [nnUNet_raw_data_base] \nnUNet_cropped_data \Task007_Pancreas 下生成了图像(.npz,包含图像和标注)和他的属性信息(.pkl),并且生成gt_segmentation保存分割的金标准
| |
结束后的文件加是
|-- nnUNet_preprocessed
|
|-- nnUNet_raw
| |
| |-- nnUNet_cropped_data
| | |
| | `-- Task007_Pancreas
| | |
| | |-- dataset.json
| | |
| | |-- gt_segmentations
| | | |-- pancreas_001.nii.gz
| | | |-- pancreas_004.nii.gz
| | | |-- ...
| | | `-- pancreas_421.nii.gz
| | |
| | |-- pancreas_001.npz
| | |-- pancreas_001.pkl
| | |-- pancreas_004.npz
| | |-- pancreas_004.pkl
| | |-- ... .npz
| | |-- ... .pkl
| | |-- pancreas_421.npz
| | `-- pancreas_421.pkl
| |
| `-- nnUNet_raw_data
| |
| `-- Task007_Pancreas
| |
| |-- dataset.json
| |
| |-- imagesTr
| | |-- pancreas_001_0000.nii.gz
| | |-- pancreas_004_0000.nii.gz
| | |-- ...
| | `-- pancreas_421_0000.nii.gz
| |
| |-- imagesTs
| | |-- pancreas_002_0000.nii.gz
| | |-- pancreas_003_0000.nii.gz
| | |-- ...
| | `-- pancreas_420_0000.nii.gz
| |
| `-- labelsTr
| |-- pancreas_001.nii.gz
| |-- pancreas_004.nii.gz
| |-- ...
| `-- pancreas_421.nii.gz
|
`-- result_folder
|
`-- nnUNet
在这个过程中可能会出现报错“ multiprocessing.pool.MaybeEncoding Error: Error sending result: ‘<multiprocessing.pool.ExceptionWithTraceback object at 0x7f8ff58e8110>’. Reason: ‘PicklingError(“Can’t pickle <class ‘MemoryError’>: it’s not the same object as builtins.MemoryError”)’”
原因是在这个过程中RAM爆了,所以多跑几次把这个跑完就好了
对数据集进行分析
| |
进行数据集分析,储存在 [nnUNet_raw_data_base]\ nnUNet_cropped_data\ Task007_Pancreas\ dataset_properties.pkl中
| |
之后将dataset_properties.pkl,还有dataset.json拷贝到了 [nnUNet_preprocessed]/Task007_Pancreas 中
对3D网络进行数据集前处理 planner_3d
nnunet $\rightarrow$ experiment_planning $\rightarrow$ experiment_planner_baseline_3DUNet.py
归一化
除CT外,其他模态使用的归一化都是z-scoringCT使用的是全局归一化. To this end, nnU-Net uses the 0.5 and 99.5 percentiles of the foreground voxels for clipping as well as the global foreground mean a standard deviation for normalization on all images
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30def determine_whether_to_use_mask_for_norm(self): # only use the nonzero mask for normalization of the cropping based on it resulted in a decrease in # image size (this is an indication that the data is something like brats/isles and then we want to # normalize in the brain region only) modalities = self.dataset_properties['modalities'] num_modalities = len(list(modalities.keys())) use_nonzero_mask_for_norm = OrderedDict() for i in range(num_modalities): if "CT" in modalities[i]: use_nonzero_mask_for_norm[i] = False else: all_size_reductions = [] for k in self.dataset_properties['size_reductions'].keys(): all_size_reductions.append(self.dataset_properties['size_reductions'][k]) if np.median(all_size_reductions) < 3 / 4.: print("using nonzero mask for normalization") use_nonzero_mask_for_norm[i] = True else: print("not using nonzero mask for normalization") use_nonzero_mask_for_norm[i] = False for c in self.list_of_cropped_npz_files: case_identifier = get_case_identifier_from_npz(c) properties = self.load_properties_of_cropped(case_identifier) properties['use_nonzero_mask_for_norm'] = use_nonzero_mask_for_norm self.save_properties_of_cropped(case_identifier, properties) use_nonzero_mask_for_normalization = use_nonzero_mask_for_norm return use_nonzero_mask_for_normalization寻找目标的CT间隔(spacings)
1 2 3 4 5 6 7 8def get_target_spacing(self): spacings = self.dataset_properties['all_spacings'] # target = np.median(np.vstack(spacings), 0) # if target spacing is very anisotropic we may want to not downsample the axis with the worst spacing # uncomment after mystery task submission # 在初始化中设定:self.target_spacing_percentile = 50 target = np.percentile(np.vstack(spacings), self.target_spacing_percentile, 0) return target将所有CT的中位数作为所有CT的目标spacing,之后利用新的spacing得到进行归一化的图像shape:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15target_spacing = self.get_target_spacing() new_shapes = [np.array(i) / target_spacing * np.array(j) for i, j in zip(spacings, sizes)] print("target_spacing:") print(target_spacing) print("sizes:") print(sizes) print("new_shapes:") print(new_shapes) # target_spacing: # [2.5, 0.80273402, 0.80273402] # sizes: # [(110, 512, 512), (107, 512, 512), ..., (131, 512, 512)] # new_shapes: # [([110., 411.09492159, 411.09492159]), ([107., 473.38251017, 473.38251017]),...,([104.8, 620.37968636, 620.37968636])]可以看到,这里图像大小归一化的策略是根据spacing对图像的所有维度进行归一化,而不是只针对axial轴进行归一化。之后他进行了spacing的大小分析:
1 2 3 4 5 6 7 8 9 10 11 12# we base our calculations on the median shape of the datasets median_shape = np.median(np.vstack(new_shapes), 0) print("the median shape of the dataset is ", median_shape) max_shape = np.max(np.vstack(new_shapes), 0) print("the max shape in the dataset is ", max_shape) min_shape = np.min(np.vstack(new_shapes), 0) print("the min shape in the dataset is ", min_shape) print("we don't want feature maps smaller than ", selfunet_featuremap_min_edge_length, " in the bottleneck") # the median shape of the dataset is [96. 512. 512.] # the max shape in the dataset is [210.28361328 622.87101959 622.87101959] # the min shape in the dataset is [51. 386.18037278 386.18037278] # we don't want feature maps smaller than 4 in the bottleneck自动计算训练策略
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20print("generating configuration for 3d_fullres") self.plans_per_stage.append(self.get_properties_for_stage( target_spacing_transposed, target_spacing_transposed, median_shape_transposed, len(self.list_of_cropped_npz_files), num_modalities, len(all_classes) + 1 ) print(self.plans_per_stage) # [{'batch_size': 2, # 'num_pool_per_axis': [3, 5, 5], # 'patch_size': array([40, 224, 224]), # 'median_patient_size_in_voxels': array([96, 512, 512]), # 'current_spacing': array([2.5, 0.80273402, 0.80273402]), # 'original_spacing': array([2.5, 0.80273402, 0.80273402]), # 'do_dummy_2D_data_aug': True, # 'pool_op_kernel_sizes': [[1, 2, 2], [2, 2, 2], [2, 2, 2], [2, 2, 2], [1, 2, 2]], # 'conv_kernel_sizes': [[1, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3]}]注意,这里的def get_properties_for_stage跳转到了experiments_planner_baseline_3DUNet.py中了
根据spacing计算,如果spacing最大的方向的体素是512,那么这个feature map的shape是多少,然后和之前选中的那个shape进行比较,选小的一边
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27""" current_spacing: [2.5 0.80273402 0.80273402] new_median_shape: [96 512 512] """ # compute how many voxels are one mm # input_path_size [0.4 1.24574265 1.24574265] input_patch_size = 1 / np.array(current_spacing) # normalize voxels per mm # 这个归一化我其实不是很懂,是不是将path等比例缩放,因为后面是1/min() # 或者我觉得更有道理的normalize应该是对mm^3进行归一化 # input_path_size [0.41501162 1.29249419 1.29249419] input_patch_size /= input_patch_size.mean() # create an isotropic patch of size 512x512x512mm # 这里的意思其实是,如果建立一个各个方向spacing都是一样的,需要的voxels,也就是map的大小, # 这里设定了最小的一个方向的大小是 512 voxel # input_path_size [ 512. 1594.55058874 1594.55058874] input_patch_size *= 1 / min(input_patch_size) * 512 # to get a starting value # input_path_size [ 512 1595 1595] input_patch_size = np.round(input_patch_size).astype(int) # clip it to the median shape of the dataset because patches larger then that make not much sense # 各个方向上取一个最小的结果 # input_path_size [ 96 512 512] input_patch_size = [min(i, j) for i, j in zip(input_patch_size, new_median_shape)]根据输入的大小,计算网络结构以及可除尽的大小
1 2 3 4 5 6 7 8 9 10 11network_num_pool_per_axis, pool_op_kernel_sizes, conv_kernel_sizes, new_shp, \ shape_must_be_divisible_by = get_pool_and_conv_props( current_spacing, input_patch_size, self.unet_featuremap_min_edge_length, self.unet_max_numpool) ## network_num_pool_per_axis: [4, 7, 7] ## pool_op_kernel_sizes: [[1, 2, 2], [2, 2, 2], [2, 2, 2], [2, 2, 2], [2, 2, 2], [1, 2, 2], [1, 2, 2]] ## conv_kernel_sizes: [[1, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3], [3, 3, 3]] ##new_shape: [ 96 512 512] ##shape_must_be_divisible_by: [ 16 128 128]自动计算网络架构:
网络中仅使用conv - >instance normalization -> Leaky relu。上采样使用的是转置卷积。确定好input_path_size后,对每个方向进行下采样,直到最终的feature map size小于 $4\times 4\times 4$
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51def get_pool_and_conv_props(spacing, patch_size, min_feature_map_size, max_numpool): """ :param spacing: :param patch_size: :param min_feature_map_size: min edge length of feature maps in bottleneck """ dim = len(spacing) current_spacing = deepcopy(list(spacing)) current_size = deepcopy(list(patch_size)) pool_op_kernel_sizes = [] conv_kernel_sizes = [] num_pool_per_axis = [0] * dim while True: # This is a problem because sometimes we have spacing 20, 50, 50 and we want to still keep pooling. # Here we would stop however. This is not what we want! Fixed in get_pool_and_conv_propsv2 min_spacing = min(current_spacing) valid_axes_for_pool = [i for i in range(dim) if current_spacing[i] / min_spacing < 2] axes = [] for a in range(dim): my_spacing = current_spacing[a] partners = [i for i in range(dim) if current_spacing[i] / my_spacing < 2 and my_spacing / current_spacing[i] < 2] if len(partners) > len(axes): axes = partners conv_kernel_size = [3 if i in axes else 1 for i in range(dim)] # exclude axes that we cannot pool further because of min_feature_map_size constraint #before = len(valid_axes_for_pool) valid_axes_for_pool = [i for i in valid_axes_for_pool if current_size[i] >= 2*min_feature_map_size] #after = len(valid_axes_for_pool) #if after == 1 and before > 1: # break valid_axes_for_pool = [i for i in valid_axes_for_pool if num_pool_per_axis[i] < max_numpool] if len(valid_axes_for_pool) == 0: break #print(current_spacing, current_size) other_axes = [i for i in range(dim) if i not in valid_axes_for_pool] pool_kernel_sizes = [0] * dim for v in valid_axes_for_pool: pool_kernel_sizes[v] = 2 num_pool_per_axis[v] += 1 current_spacing[v] *= 2 current_size[v] = np.ceil(current_size[v] / 2) for nv in other_axes: pool_kernel_sizes[nv] = 1 pool_op_kernel_sizes.append(pool_kernel_sizes) conv_kernel_sizes.append(conv_kernel_size) #print(conv_kernel_sizes) must_be_divisible_by = get_shape_must_be_divisible_by(num_pool_per_axis) patch_size = pad_shape(patch_size, must_be_divisible_by) # we need to add one more conv_kernel_size for the bottleneck. We always use 3x3(x3) conv here conv_kernel_sizes.append([3]*dim) return num_pool_per_axis, pool_op_kernel_sizes, conv_kernel_sizes, patch_size, must_be_divisible_by实施预处理
nnunet $\rightarrow$ preprocessing $\rightarrow$ preprocessing.py $\rightarrow$ class GenericPreprocessor
主要运行函数是def _run_internal
加载cropped数据,之后进行resample
这里的resample是根据target spacing对数据和标签进行重采样到指定的spacing
然后进行数值的调整,clip掉$HU \in [… ,0.5%] & [99.5%, …]$
然后对HU进行z-score归一化
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82def resample_and_normalize(self, data, target_spacing, properties, seg=None, force_separate_z=None): """ data and seg must already have been transposed by transpose_forward. properties are the un-transposed values (spacing etc) :param data: :param target_spacing: :param properties: :param seg: :param force_separate_z: :return: """ # target_spacing is already transposed, properties["original_spacing"] is not so we need to transpose it! # data, seg are already transposed. Double check this using the properties original_spacing_transposed = np.array(properties["original_spacing"])[self.transpose_forward] before = { 'spacing': properties["original_spacing"], 'spacing_transposed': original_spacing_transposed, 'data.shape (data is transposed)': data.shape } # remove nans data[np.isnan(data)] = 0 print('data shape: {}'.format(data.shape)) data, seg = resample_patient(data, seg, np.array(original_spacing_transposed), target_spacing, 3, 1, force_separate_z=force_separate_z, order_z_data=0, order_z_seg=0, separate_z_anisotropy_threshold=self.resample_separate_z_anisotropy_threshold) after = { 'spacing': target_spacing, 'data.shape (data is resampled)': data.shape } print("before:", before, "\nafter: ", after, "\n") if seg is not None: # hippocampus 243 has one voxel with -2 as label. wtf? seg[seg < -1] = 0 properties["size_after_resampling"] = data[0].shape properties["spacing_after_resampling"] = target_spacing use_nonzero_mask = self.use_nonzero_mask assert len(self.normalization_scheme_per_modality) == len(data), "self.normalization_scheme_per_modality " \ "must have as many entries as data has " \ "modalities" assert len(self.use_nonzero_mask) == len(data), "self.use_nonzero_mask must have as many entries as data" \ " has modalities" for c in range(len(data)): scheme = self.normalization_scheme_per_modality[c] if scheme == "CT": # clip to lb and ub from train data foreground and use foreground mn and sd from training data assert self.intensityproperties is not None, "ERROR: if there is a CT then we need intensity properties" mean_intensity = self.intensityproperties[c]['mean'] std_intensity = self.intensityproperties[c]['sd'] lower_bound = self.intensityproperties[c]['percentile_00_5'] upper_bound = self.intensityproperties[c]['percentile_99_5'] data[c] = np.clip(data[c], lower_bound, upper_bound) data[c] = (data[c] - mean_intensity) / std_intensity if use_nonzero_mask[c]: data[c][seg[-1] < 0] = 0 elif scheme == "CT2": # clip to lb and ub from train data foreground, use mn and sd form each case for normalization assert self.intensityproperties is not None, "ERROR: if there is a CT then we need intensity properties" lower_bound = self.intensityproperties[c]['percentile_00_5'] upper_bound = self.intensityproperties[c]['percentile_99_5'] mask = (data[c] > lower_bound) & (data[c] < upper_bound) data[c] = np.clip(data[c], lower_bound, upper_bound) mn = data[c][mask].mean() sd = data[c][mask].std() data[c] = (data[c] - mn) / sd if use_nonzero_mask[c]: data[c][seg[-1] < 0] = 0 else: if use_nonzero_mask[c]: mask = seg[-1] >= 0 else: mask = np.ones(seg.shape[1:], dtype=bool) data[c][mask] = (data[c][mask] - data[c][mask].mean()) / (data[c][mask].std() + 1e-8) data[c][mask == 0] = 0 return data, seg, properties标签随机选点,平衡标签
这种平衡标签的方式真的是特别amazing,之前从来没想过可以这么平衡标签
他把label中的所有坐标点提取出来,然后进行随机的选点,设定随机的点最多10000个,最小不能少于标签坐标的1%。
这种的话就不需要对图像进行裁切来进行平衡,如果有斑驳的话也可以使用后期的出来来进行弥补最后data[‘class_locations’]就是[num,3]的数组,保存了选择的label的点
但是这个10000的选择不知道适不适用于其他的任何数据集???
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22# we need to find out where the classes are and sample some random locations # let's do 10.000 samples per class # seed this for reproducibility! num_samples = 10000 min_percent_coverage = 0.01 # at least 1% of the class voxels need to be selected, otherwise it may be too sparse rndst = np.random.RandomState(1234) class_locs = {} for c in all_classes: all_locs = np.argwhere(all_data[-1] == c) if len(all_locs) == 0: class_locs[c] = [] continue target_num_samples = min(num_samples, len(all_locs)) target_num_samples = max(target_num_samples, int(np.ceil(len(all_locs) * min_percent_coverage))) selected = all_locs[rndst.choice(len(all_locs), target_num_samples, replace=False)] class_locs[c] = selected print(c, target_num_samples) properties['class_locations'] = class_locs两阶段Cascade Unet
第一阶段unet是小图像 [107, 225, 225],第二阶段是全图像 [110, 411, 411]。
但是每个阶段的图像都是全景图像。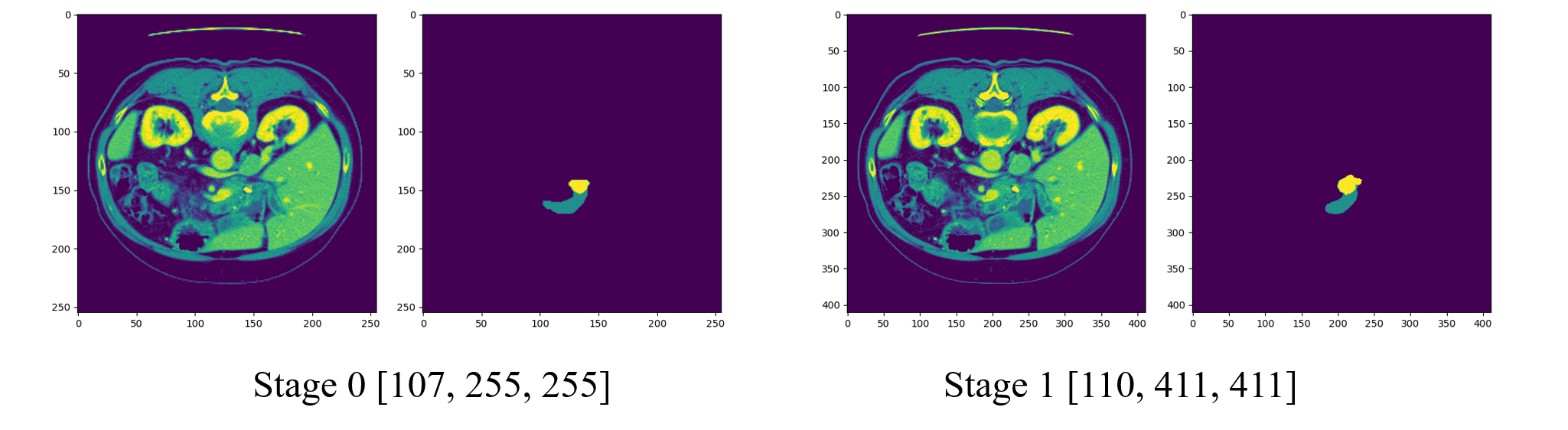
进行训练
使用 3D U-Net Cascade 进行训练
这一部分的代码在 nnunet -> run -> run_training.py 中
3D low resolution U-Net
For FOLD in [0, 1, 2, 3, 4], run:1 2 3nnUNet_train 3d_lowres nnUNetTrainerV2 TaskXXX_MYTASK FOLD --npz ## Example nnUNet_train 3d_lowres nnUNetTrainerV2 Task007_Pancreas 0 --npz3D full resolution U-Net
For FOLD in [0, 1, 2, 3, 4], run:1 2 3nnUNet_train 3d_cascade_fullres nnUNetTrainerV2CascadeFullRes TaskXXX_MYTASK FOLD --npz ## Example nnUNet_train 3d_cascade_fullres nnUNetTrainerV2CascadeFullRes Task007_Pancreas 0 --npz
损失函数
nnunet -> training -> loss_function -> dice_loss.py
| |
使用方法:
| |
如果是多个标签,会使用权重进行分配,在MultiOutputLoss2中
| |
生成训练数据集
nnunet -> training -> network_training -> nnUNetTrainerV2.py
| |
nnunet -> training -> network_training -> nnUNetTrainer.py
| |
nnunet -> training -> dataloading -> dataloading.py -> def load_dataset()
| |
nnunet -> training -> dataloading -> dataloading.py -> class DataLoader3D()
| |
学习率初始是0.01
进行了梯度剪裁
1torch.nn.utils.clip_grad_norm_(self.network.parameters(), 12)网络当中还是随机裁剪了,进入网络path的大小为(2,1,64,192,192)
图像为原尺寸大小,但是label被缩小成了每一层的大小。注意,这里的label包含了所有的标签,同时包含了胰腺和肿瘤
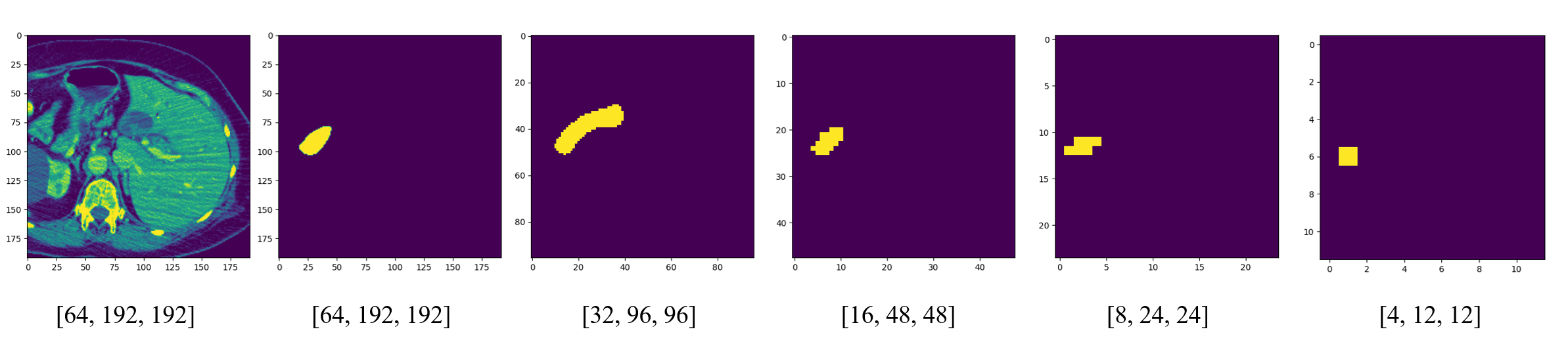
网络也会同时输出五层的结果,在loss中通过权重进行分配,权重分别是体素的比
$$\begin{aligned}64 &\times &192 &\times &192 &(64) &: \32 &\times &96 &\times &96 &(32) &: \16 &\times &48 &\times &48 &(16) &: \8 &\times &24 &\times &24 &(8) &: \4 &\times &12 &\times &12 &(1) &:\end{aligned}$$
更改(windows)
- maybe_mkdir_p $\rightarrow$ os.makedirs